
I have zero biology credentials. In 2 hours, I designed an immune-cardiac organoid protocol for Opentrons Flex that Perplexity's deep research called "scientifically mature" across 108 sources, after I told it to roast the idea as "freshman-level."
Either the AI is lying, or something fundamental just changed in how workflows can get built.
This year, we’ve hosted two more community hacks showing how AI for science changes how fast things can get built, designed, and tested. The first one was around building Physical MCPs, and the second was a 24hr cell cultivation challenge.

These three agent skills came out of a community workshop with Opentrons as we wrapped the year. Here’s what got built:
Literature review → to understand interesting ideas
Hypothesis generation → to generate direction for experiment
Protocol designer → a Python script that Opentrons Flex can use
Claude Code debugs the errors. The Opentrons simulator validates. Then, Perplexity deep research as LLM-as-judge to gut-check the output.
The verdict's still out on whether this is real or AI slop. There’s more work to be done on the meta-systems for designing and testing nodes in a workflow.
Everything is open source on GitHub if you want to try it yourself.
First, What Are Agent Skills by Anthropic?

Last year, Anthropic released Model Context Protocol (MCP), which are connectors that let LLMs use tools like Notion, GitHub, Stripe, and thousands more. This year, they released a way to give LLMs domain knowledge and workflows called Agent Skills.
If LLMs were cooks in a kitchen, MCPs would be the tools and appliances like a knife, pot, and oven, and skills are the recipes that tell the LLM how to use them together to cook up specific dishes. Just like a recipe can be written to have different guidelines, steps, and substeps, skills help capture specific perspectives to get better outputs than a single LLM call.
A skill has three parts:
YAML header: 2 lines loaded into every LLM call so Claude knows what's available and can decide when to use a tool
SKILL.md: instructions, guidelines, and reference material in markdown
Supporting files: examples, scripts, even nested MCPs
The wild part: one of the first skills Anthropic released was Skill Creator. Claude can build its own skills.
Here’s what that looks like for Skills Creator skill:
YAML Header
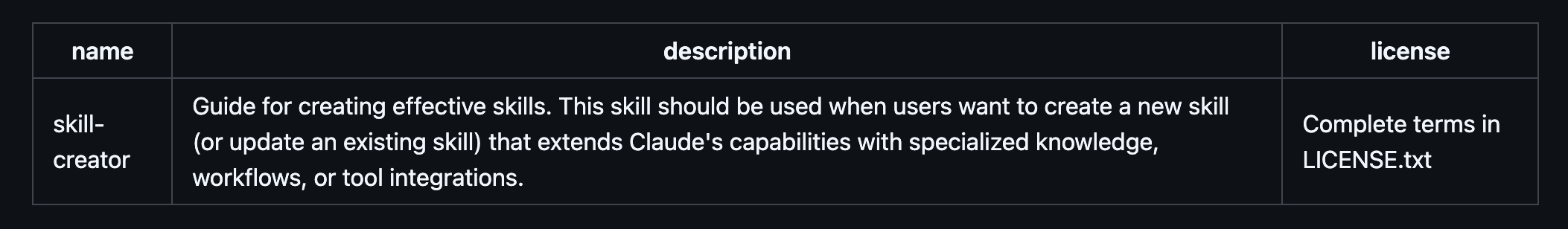
Selected sections of skill-creator.md
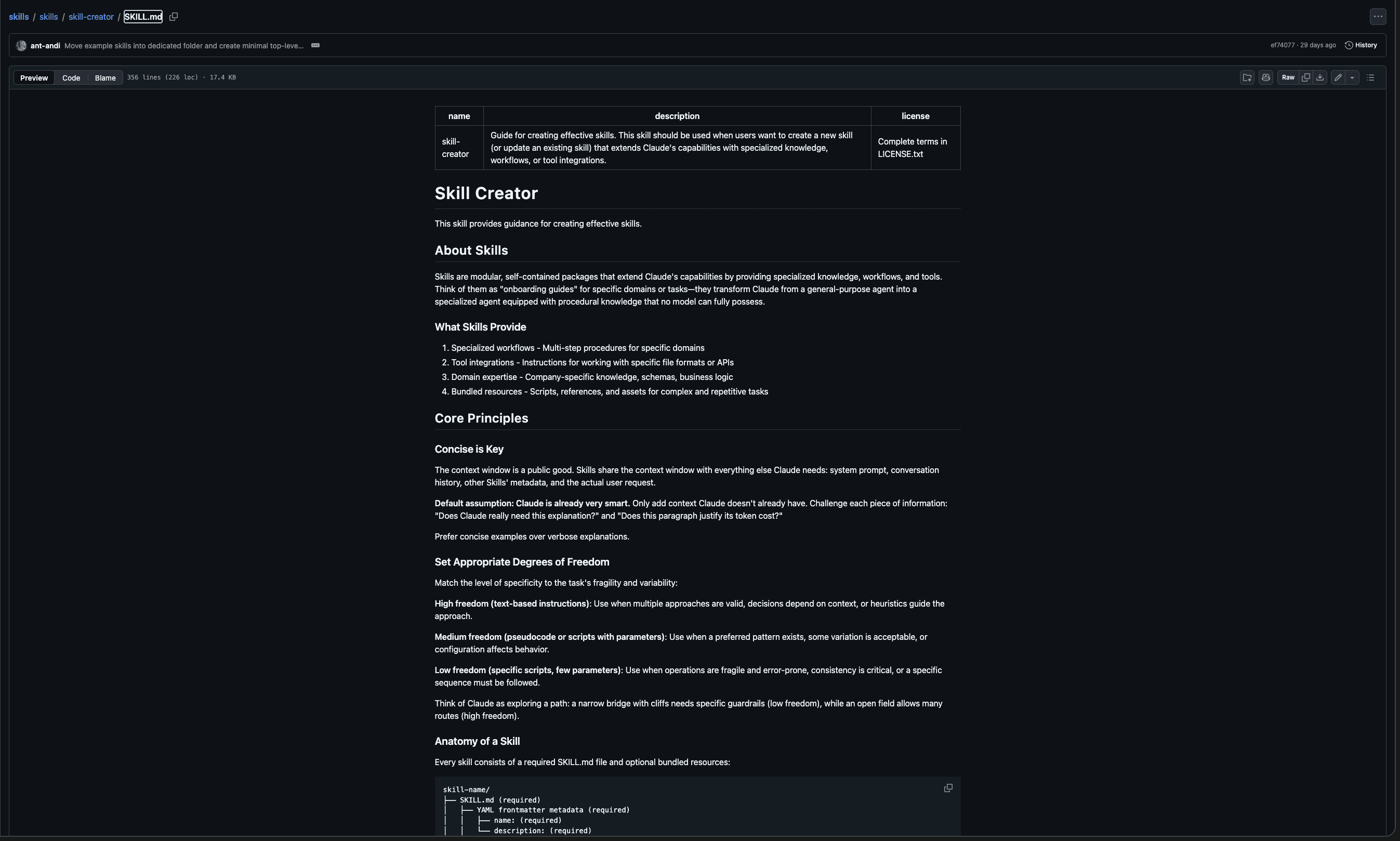
Files included in skill creator include output-patterns.md and workflows.md as references and also init_skill.py as a template and package_skill.py
Here’s the link to Skill Creator on GitHub.
And to install a skill, it’s as easy as uploading a ZIP file through Claude Desktop app. Once you start making skills with Skills Creator, they can save with one button and shared easily.
Skills aren’t just an Anthropic thing by Claude. They are being adopted by Open AI and other model providers as a standard way to make LLMs work better for specific tasks.
From Skills Creator to 3 Life Science Skills Created
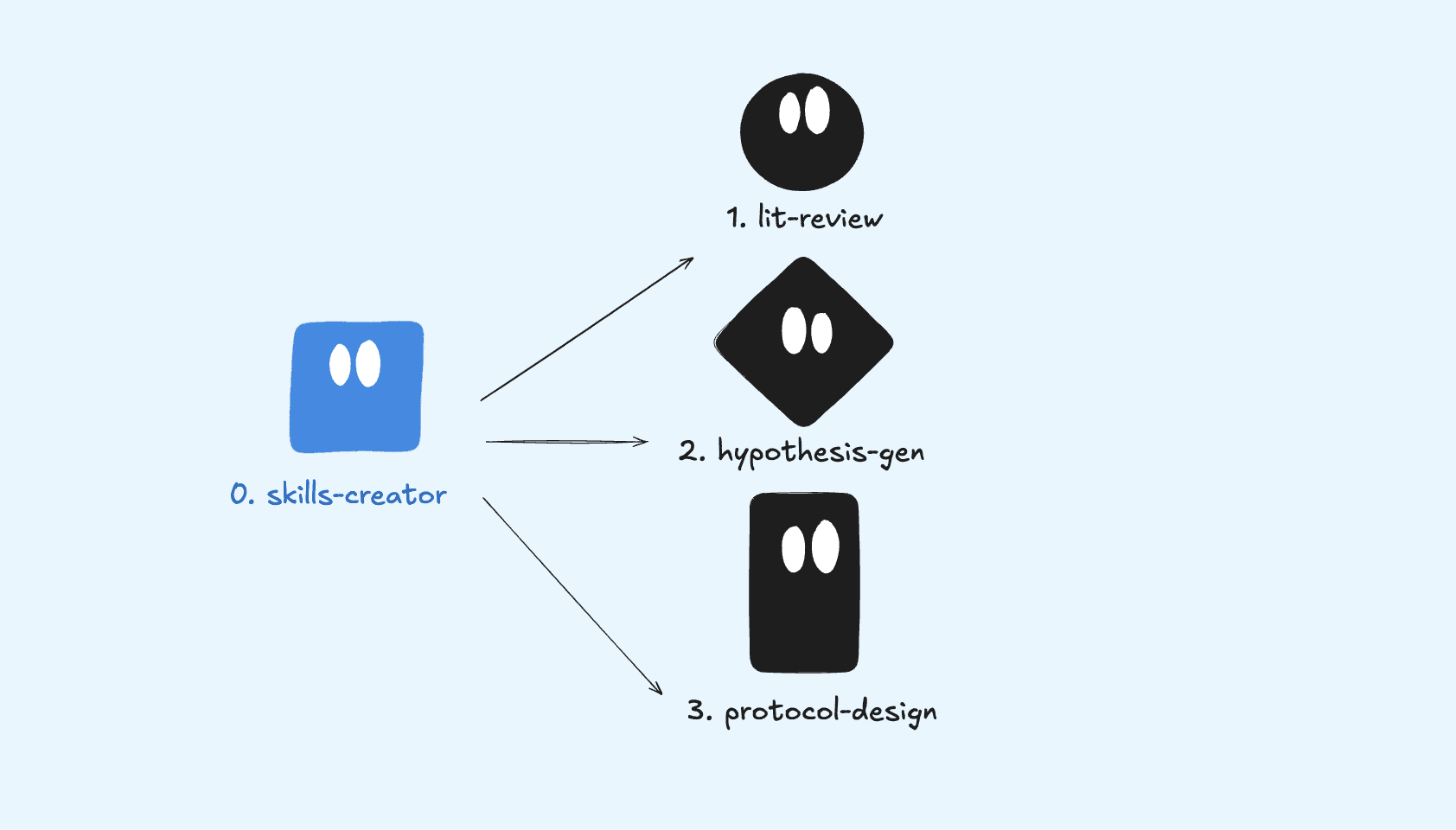
I used Skills Creator to build an agentic workflow that goes from research question to executable Opentrons protocol. I picked cardiac organoids because it's specific enough to stress test the system. This also acts as v1 tooling for a microfluidics hack we’re planning in 2026.
You can download and run these skills for yourself.
The system has three skills that chain together like a pipeline:
Let’s break each one down.
The Workflow in Action
Step 1: Start the workflow in natural language
Prompt:
Right now, it’s not clear when Claude chooses what skills to use, so for this one, I made it more explicit in the first ask.
What came back:
The skill identified that the immune-cardiac axis is a critical gap in organoid research. Here’s sample:
Here's why:
Cardiac resident macrophages are absent from nearly all current organoid models
These macrophages eliminate dysfunctional mitochondria from cardiomyocytes
They regulate cardiac conduction via gap junctions
They mediate fibrosis and inflammation post-injury
Current models either have mature cells but no structure (Mills), or chambers but immature cells (Mendjan). Nobody has integrated immune components.
Full output from Claude Skills here.
Step 2: It took ideas to generate hypotheses
Prompt: Nothing new was needed, as it asked me if I wanted to continue to second step.
What came back (top 4):
Rank | Hypothesis | Score |
|---|---|---|
1 | Inflammatory cytokine (TNF-α/IL-1β) effects on calcium dynamics | 4.4/5.0 |
2 | Macrophage-conditioned media effects on beating | 4.1/5.0 |
3 | Doxorubicin dose-response (classic cardiotox) | 3.9/5.0 |
4 | AMPK maturation cocktail acute effects | 3.5/5.0 |
The top hypothesis won because it’s novel (immune-cardiac axis in organoids is unexplored), feasible (uses pre-made organoids and endpoint calcium imaging), and has clear clinical relevance (heart failure, myocarditis).
Step 3: Generate the Protocol
What came out: A complete Python protocol with plate layout, reagent guide, and 72 wells treated across 6 conditions.
The Code, Line by Line
Here's the protocol, with explanations for each section. This is the stuff that actually runs on the robot.
The Metadata
Every Opentrons protocol starts with metadata. Here's the critical part:
Key point: For Opentrons Flex, apiLevel goes in requirements, NOT in metadata. This tripped up the first version. The OT-2 (older robot) puts it in metadata. Flex requires it in requirements.
Labware Setup
Key point: The trash bin line is required for Flex. The OT-2 has a built-in fixed trash. Flex makes you define it explicitly. Without this line, you get a runtime error.
The Critical Pipette Selection
This is where the first two versions failed. Here's why:
Why does this matter?
An 8-channel pipette dispenses to ALL 8 ROWS (A-H) simultaneously. When you target plate['A1'] with an 8-channel, it actually dispenses to A1, B1, C1, D1, E1, F1, G1, H1.
Look at our plate layout:
Rows A-B get different treatments than rows C-D. If we used an 8-channel targeting column 1, we'd put the same reagent in rows A through H of that column. That's wrong. Different rows need different treatments.
The rule:
Scattered individual wells → single-channel
Full columns (all A-H same) → 8-channel
Full plate (all 96 same) → 96-channel
Running the Protocol
After treatments, there's a 24-hour incubation. The protocol pauses and waits for you to press resume. Then it adds Fluo-4 calcium dye to all 72 wells for the imaging readout.
The Bugs and How They Got Fixed
The first two protocol versions failed. Here's what went wrong:
Bug | Why It Broke | The Fix |
|---|---|---|
8-channel with scattered wells | Dispenses to entire column A-H | Use single-channel |
Duplicate apiLevel | In both metadata AND requirements | Keep only in requirements |
Missing trash bin | Flex requires explicit trash | Add |
How we found them:
In the true spirit of vibe designing this experiment, I let Claude code debug errors coming from the Opentrons simulator:
The errors are pretty clear:
"Cannot create a protocol with api level X in metadata and requirements"
"No trash container defined"
Claude Code read the error messages, looked at the skill's reference files documenting the correct patterns, and fixed the issues.
The third version passes simulation and is ready for hardware. That protocol is here.
LLM as Judge: Is This Real Science or AI Slop?
This is the main question I actually care about.
Did Claude just make this up?
Is the hypothesis scientifically valid?
I used Perplexity AI idea labs, selecting academic resources, as one independent judge. I also negatively biased the evaluation by saying it’s ‘freshman’ level work.
The prompt:
What came back (from 108+ sources):
Perplexity could be wrong. LLMs can confidently cite things that don't exist. But it provided 108 source URLs, many from recent papers in Nature, Cell Death & Disease, and eLife.
The logic chain checks out: if cytokines disrupt calcium in 2D cardiomyocytes (established), and organoids are the next-generation model (established), then testing cytokines in organoids is a reasonable hypothesis (valid).
Is this proof that it should get a ton of funding for being a potential breakthrough? No.
Is it a good signal that I didn't waste 3 hours generating nonsense? Yes.
Here’s Perplexity’s full output.
There’s more work to be done here to evaluate individual steps as well as final output.
How to Try This Yourself
Step 1: Get the Skills
Upload each .zip file to Claude's Skills interface in settings on Claude Desktop.
Step 2: Run the Workflow
Claude has a bias towards using all of the skills.
Step 3: Test with the Simulator
You'll need Python 3.10+:
If it runs without errors, it's ready for hardware.
Step 4: Adapt for Your Research
Fork the repo. The skills are modular. The key files to edit:
references/field-landscape.md— swap in your field's major labsreferences/hackathon-constraints.md— update with your equipment and timelinereferences/flex-specifications.md— already complete for Opentrons Flex
Build your own domain-specific skills.
The Point of All This
Let me answer the question I started with: is this valuable science, or is this AI slop?
The honest answer: somewhere in between, and that's the point.
What's different now:
Then (1 year ago) | Now |
|---|---|
Took a Google L6 engineer to spin up multiple agents to run this flow | 3 hours on a Thursday night makes a custom 3 step workflow from scratch |
Required domain expertise to even start | Skills encode domain knowledge which can be further iterated and designed |
Protocols had to be written from scratch | Templates + validation built in |
What hasn't changed:
The human still has to validate
The human still has to test on hardware
The human still has to interpret results
We need better evals to see what matters
Skills don't replace expertise. They encode it. They make the activation energy lower. They let someone who doesn't have the PhD at least get to a starting point that makes sense.
The reference files in these skills, the field landscape, the methodology guides, the specifications, that's the knowledge that used to be locked in specific institutions and individuals. Now it's composable, remixable, shareable, and improvable.
And that seems like a wild start to 2026.
ICYMI: Here’s the GitHub Repository. And here are more science based skills by K-Dense.
Thank you to our partners!
Luis and the Bay Area Lab Automators for co-hosting builds in the community
Carter and the Monomer Bio team for the community support for builds
Shanin, Homam, Krishna, and the whole Opentrons team for supporting builds like this
About the writer
Michael Raspuzzi is founder of Worldwide Studios, where he enables people to build with the latest across AI, robotics, and applied science in community hacks and programs.
Claude Opus 4.5 through Claude Code is like a supercharged associate helping with the draft of this. It still doesn’t quite automate the writing, but it augments the process well.
Next, we’re exploring how to design better skills and also evals to judge output from each skill. If you want to chat, reach out. The best part of building in public is the conversations that come after.